Repository
» Data AccessID 108 » Model AccessID 44
Detail View - Model AccessID 44
General Information

| Manuscript title | Kinetic modeling of tricarboxylic acid cycle and glyoxylate bypass in Mycobacterium tuberculosis, and its application to assessment of drug targets |
| PubMed ID | 16887020 |
| Journal | Theoretical Biology and Medical Modelling |
| Year | 2006 |
| Authors | Vivek Kumar Singh and Indira Ghosh |
| Affiliations | Bioinformatics Centre, University of Pune, India |
| Keywords | Tuberculosis, Steady State Flux, Glyoxylate Bypass, Metabolic Control Analysis, Experimental Flux |
| Full text article |
 Singh_2006.pdf
Singh_2006.pdf
|
| Project name | not specified |
| Data used for | Model validation |
Model Information

| Model name | Singh2006_TCA_Ecoli_glucose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Escherichia coli | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Model type | ordinary differential equations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Category | Metabolism | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of reactions | 11 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of species | 12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of regulators | not specified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of parameters | 45 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of compartments | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dilution rate (h-1) | — | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Model file(s) and history |
 LATEST BIOMD0000000222.xml
(2018-07-23 10:39:44 UTC
by Administrator KiMoSysFirst name: Administrator
LATEST BIOMD0000000222.xml
(2018-07-23 10:39:44 UTC
by Administrator KiMoSysFirst name: AdministratorAffiliation: INESC-ID/IST Homepage: http://kdbio.inesc-id.pt/kimosys Interests: mathematical modeling, accessible data, use of data )
version comment: model for glucose condition
Preview
Preview file BIOMD0000000222.xml<?xml version='1.0' encoding='UTF-8' standalone='no'?>
<!-- This model was downloaded from BioModels Database -->
<!-- http://www.ebi.ac.uk/biomodels-static/ -->
<!-- Mon Jul 23 11:06:13 BST 2018 -->
<sbml level='2' metaid='metaid_0000001' version='4' xmlns='http://www.sbml.org/sbml/level2/version4'>
<model id='Singh2006_TCA_Ecoli_glucose' metaid='_061956' name='Singh2006_TCA_Ecoli_glucose'>
<notes>
<body xmlns='http://www.w3.org/1999/xhtml'>
<p>This a model from the article: <br/>
<strong>Kinetic modeling of tricarboxylic acid cycle and glyoxylate bypass in Mycobacterium tuberculosis, and its application to assessment of drugtargets.</strong>
<br/>
Singh VK , Ghosh I <em>Theor Biol Med Model</em>
2006 Aug 3;3:27 <a href='http://www.ncbi.nlm.nih.gov/pubmed/16887020'>16887020</a>
, <br/>
<strong>Abstract:</strong>
<br/>
BACKGROUND: Targeting persistent tubercule bacilli has become an important challenge in the development of anti-tuberculous drugs. As the glyoxylate bypass is essential for persistent bacilli, interference with it holds the potential for designing new antibacterial drugs. We have developed kinetic models of the tricarboxylic acid cycle and glyoxylate bypass in Escherichia coli and Mycobacterium tuberculosis, and studied the effects of inhibition of various enzymes in the M. tuberculosis model. RESULTS: We used E. coli to validate the pathway-modeling protocol and showed that changes in metabolic flux can be estimated from gene expression data. The M. tuberculosis model reproduced the observation that deletion of one of the two isocitrate lyase genes has little effect on bacterial growth in macrophages, but deletion of both genes leads to the elimination of the bacilli from the lungs. It also substantiated the inhibition of isocitrate lyases by 3-nitropropionate. On the basis of our simulation studies, we propose that: (i) fractional inactivation of both isocitrate dehydrogenase 1 and isocitrate dehydrogenase 2 is required for a flux through the glyoxylate bypass in persistent mycobacteria; and (ii) increasing the amountof active isocitrate dehydrogenases can stop the flux through the glyoxylate bypass, so the kinase that inactivates isocitrate dehydrogenase 1 and/or the proposed inactivator of isocitrate dehydrogenase 2 is a potential target for drugs against persistent mycobacteria. In addition, competitive inhibition of isocitrate lyases along with a reduction in the inactivation of isocitrate dehydrogenases appears to be a feasible strategy for targeting persistent mycobacteria. CONCLUSION: We used kinetic modeling of biochemical pathways to assess various potential anti-tuberculous drug targets that interfere with the glyoxylate bypass flux, and indicated the type of inhibition needed to eliminate the pathogen. The advantage of such an approach to the assessment of drug targets is that it facilitates the study of systemic effect(s) of the modulation of the target enzyme(s) in the cellular environment. </p>
<p>This model originates from BioModels Database: A Database of Annotated Published Models (http://www.ebi.ac.uk/biomodels/). It is copyright (c) 2005-2010 The BioModels.net Team. <br/>
For more information see the <a href='http://www.ebi.ac.uk/biomodels/legal.html' target='_blank'>terms of use</a>
. <br/>
To cite BioModels Database, please use: <a href='http://www.ncbi.nlm.nih.gov/pubmed/20587024' target='_blank'>Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, Snoep JL, Hucka M, Le Novère N, Laibe C (2010) BioModels Database: An enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol., 4:92.</a>
</p>
</body>
</notes>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:dc='http://purl.org/dc/elements/1.1/' xmlns:dcterms='http://purl.org/dc/terms/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#' xmlns:vCard='http://www.w3.org/2001/vcard-rdf/3.0#'>
<rdf:Description rdf:about='#_061956'>
<dc:creator>
<rdf:Bag>
<rdf:li rdf:parseType='Resource'>
<vCard:N rdf:parseType='Resource'>
<vCard:Family>Chelliah</vCard:Family>
<vCard:Given>Vijayalakshmi</vCard:Given>
</vCard:N>
<vCard:EMAIL>viji@ebi.ac.uk</vCard:EMAIL>
<vCard:ORG rdf:parseType='Resource'>
<vCard:Orgname>EMBL-EBI</vCard:Orgname>
</vCard:ORG>
</rdf:li>
<rdf:li rdf:parseType='Resource'>
<vCard:N rdf:parseType='Resource'>
<vCard:Family>Singh</vCard:Family>
<vCard:Given>Vivek Kumar</vCard:Given>
</vCard:N>
<vCard:EMAIL>vivek@bioinfo.ernet.in</vCard:EMAIL>
<vCard:ORG rdf:parseType='Resource'>
<vCard:Orgname>University of Pune</vCard:Orgname>
</vCard:ORG>
</rdf:li>
</rdf:Bag>
</dc:creator>
<dcterms:created rdf:parseType='Resource'>
<dcterms:W3CDTF>2006-09-29T22:47:42Z</dcterms:W3CDTF>
</dcterms:created>
<dcterms:modified rdf:parseType='Resource'>
<dcterms:W3CDTF>2010-12-20T09:47:18Z</dcterms:W3CDTF>
</dcterms:modified>
<bqmodel:is>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/biomodels.db/MODEL8583955822'/>
</rdf:Bag>
</bqmodel:is>
<bqmodel:is>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/biomodels.db/BIOMD0000000222'/>
</rdf:Bag>
</bqmodel:is>
<bqmodel:isDescribedBy>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/pubmed/16887020'/>
</rdf:Bag>
</bqmodel:isDescribedBy>
<bqbiol:hasTaxon>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/taxonomy/562'/>
</rdf:Bag>
</bqbiol:hasTaxon>
<bqbiol:isHomologTo>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/reactome/REACT_1785'/>
</rdf:Bag>
</bqbiol:isHomologTo>
<bqbiol:hasVersion>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.go/GO:0006097'/>
<rdf:li rdf:resource='http://identifiers.org/obo.go/GO:0006099'/>
</rdf:Bag>
</bqbiol:hasVersion>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.pathway/ko00020'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfUnitDefinitions>
<unitDefinition id='time' metaid='_871153' name='min'>
<listOfUnits>
<unit kind='second' metaid='_871165' multiplier='60'/>
</listOfUnits>
</unitDefinition>
<unitDefinition id='substance' metaid='_871177' name='mmole'>
<listOfUnits>
<unit kind='mole' metaid='_871189' scale='-3'/>
</listOfUnits>
</unitDefinition>
<unitDefinition id='mmlmin' metaid='metaid_0000031' name='mM_per_min'>
<listOfUnits>
<unit kind='mole' metaid='_871201' scale='-3'/>
<unit exponent='-1' kind='litre' metaid='_871214'/>
<unit exponent='-1' kind='second' metaid='_871226' multiplier='60'/>
</listOfUnits>
</unitDefinition>
<unitDefinition id='mml' metaid='metaid_0000032' name='mM'>
<listOfUnits>
<unit kind='mole' metaid='_871239' scale='-3'/>
<unit exponent='-1' kind='litre' metaid='_871251'/>
</listOfUnits>
</unitDefinition>
</listOfUnitDefinitions>
<listOfCompartments>
<compartment id='cell' metaid='_061967' size='1'/>
</listOfCompartments>
<listOfSpecies>
<species boundaryCondition='true' compartment='cell' id='aca' initialConcentration='0.5' metaid='_061987' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_061987'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:15351'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00024'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species boundaryCondition='true' compartment='cell' id='oaa' initialConcentration='0.004' metaid='_062007' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062007'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:30744'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00036'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species boundaryCondition='true' compartment='cell' id='coa' initialConcentration='0.0001' metaid='_062027' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062027'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:15346'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C000010'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='cit' initialConcentration='3' metaid='_062047' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062047'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:30769'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00158'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='icit' initialConcentration='0.018' metaid='_062067' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062067'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:30887'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00311'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='akg' initialConcentration='0.2' metaid='_062087' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062087'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:30915'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00026'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='sca' initialConcentration='0.04' metaid='_062107' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062107'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:15380'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00091'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='suc' initialConcentration='0.6' metaid='_062127' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062127'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:15741'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00042'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='fa' initialConcentration='0.3' metaid='_062147' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062147'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:18012'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00122'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='mal' initialConcentration='1.8' metaid='_062167' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062167'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:6650'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00711'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species compartment='cell' id='gly' initialConcentration='4' metaid='_062187' sboTerm='SBO:0000247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062187'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.chebi/CHEBI:16891'/>
<rdf:li rdf:resource='http://identifiers.org/kegg.compound/C00048'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
</species>
<species boundaryCondition='true' compartment='cell' id='biosyn' initialConcentration='0.1' metaid='_062207'/>
</listOfSpecies>
<listOfReactions>
<reaction id='CS' metaid='_062227'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062227'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00351'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871263' species='aca'/>
<speciesReference metaid='_871275' species='oaa'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871287' species='coa'/>
<speciesReference metaid='_871299' species='cit'/>
</listOfProducts>
<kineticLaw metaid='_871311'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_cs </ci>
<apply>
<divide/>
<ci> aca </ci>
<ci> Kaca_cs </ci>
</apply>
<apply>
<divide/>
<ci> oaa </ci>
<ci> Koaa_cs </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_cs </ci>
<apply>
<divide/>
<ci> coa </ci>
<ci> Kcoa_cs </ci>
</apply>
<apply>
<divide/>
<ci> cit </ci>
<ci> Kcit_cs </ci>
</apply>
</apply>
</apply>
<apply>
<times/>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> aca </ci>
<ci> Kaca_cs </ci>
</apply>
<apply>
<divide/>
<ci> coa </ci>
<ci> Kcoa_cs </ci>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> oaa </ci>
<ci> Koaa_cs </ci>
</apply>
<apply>
<divide/>
<ci> cit </ci>
<ci> Kcit_cs </ci>
</apply>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_cs' metaid='_287404' sboTerm='SBO:0000350' units='mmlmin' value='91.2'/>
<parameter id='Kaca_cs' metaid='_287405' sboTerm='SBO:0000322' units='mml' value='0.03'/>
<parameter id='Koaa_cs' metaid='_287406' sboTerm='SBO:0000322' units='mml' value='0.07'/>
<parameter id='Vr_cs' metaid='_287408' sboTerm='SBO:0000353' units='mmlmin' value='0.912'/>
<parameter id='Kcoa_cs' metaid='_287409' sboTerm='SBO:0000323' units='mml' value='0.3'/>
<parameter id='Kcit_cs' metaid='_287410' sboTerm='SBO:0000323' units='mml' value='0.7'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='ACN' metaid='_062247'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062247'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R01324'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871323' species='cit'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871335' species='icit'/>
</listOfProducts>
<kineticLaw metaid='_871348'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_acn </ci>
<apply>
<divide/>
<ci> cit </ci>
<ci> Kcit_acn </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_acn </ci>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_acn </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> cit </ci>
<ci> Kcit_acn </ci>
</apply>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_acn </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_acn' metaid='_287411' sboTerm='SBO:0000350' units='mmlmin' value='91.2'/>
<parameter id='Kcit_acn' metaid='_287412' sboTerm='SBO:0000322' units='mml' value='1.7'/>
<parameter id='Vr_acn' metaid='_287413' sboTerm='SBO:0000353' units='mmlmin' value='0.912'/>
<parameter id='Kicit_acn' metaid='_287414' sboTerm='SBO:0000323' units='mml' value='3.33'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='ICD' metaid='_062267'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062267'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00709'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871360' species='icit'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871372' species='akg'/>
</listOfProducts>
<kineticLaw metaid='_871385'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_icd </ci>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icd </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_icd </ci>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_icd </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icd </ci>
</apply>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_icd </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_icd' metaid='_287415' sboTerm='SBO:0000350' units='mmlmin' value='14.72'/>
<parameter id='Kicit_icd' metaid='_287416' sboTerm='SBO:0000322' units='mml' value='0.008'/>
<parameter id='Vr_icd' metaid='_287417' sboTerm='SBO:0000353' units='mmlmin' value='0.1472'/>
<parameter id='Kakg_icd' metaid='_287419' sboTerm='SBO:0000323' units='mml' value='0.13'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='KDH' metaid='_062287'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062287'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R08549'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871397' species='akg'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871409' species='sca'/>
</listOfProducts>
<kineticLaw metaid='_871421'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_kdh </ci>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_kdh </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_kdh </ci>
<apply>
<divide/>
<ci> sca </ci>
<ci> Ksca_kdh </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_kdh </ci>
</apply>
<apply>
<divide/>
<ci> sca </ci>
<ci> Ksca_kdh </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_kdh' metaid='_287421' sboTerm='SBO:0000350' units='mmlmin' value='35.84'/>
<parameter id='Kakg_kdh' metaid='_287422' sboTerm='SBO:0000322' units='mml' value='0.1'/>
<parameter id='Vr_kdh' metaid='_287424' sboTerm='SBO:0000353' units='mmlmin' value='0.3584'/>
<parameter id='Ksca_kdh' metaid='_287425' sboTerm='SBO:0000323' units='mml' value='1'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='ScAS' metaid='_062307'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062307'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00407'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871433' species='sca'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871445' species='suc'/>
</listOfProducts>
<kineticLaw metaid='_871457'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_scas </ci>
<apply>
<divide/>
<ci> sca </ci>
<ci> Ksca_scas </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_scas </ci>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_scas </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> sca </ci>
<ci> Ksca_scas </ci>
</apply>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_scas </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_scas' metaid='_287427' sboTerm='SBO:0000350' units='mmlmin' value='3.5'/>
<parameter id='Ksca_scas' metaid='_287428' sboTerm='SBO:0000322' units='mml' value='0.02'/>
<parameter id='Vr_scas' metaid='_287430' sboTerm='SBO:0000353' units='mmlmin' value='0.035'/>
<parameter id='Ksuc_scas' metaid='_287431' sboTerm='SBO:0000323' units='mml' value='5'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='SDH' metaid='_062327'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062327'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00402'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871469' species='suc'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871481' species='fa'/>
</listOfProducts>
<kineticLaw metaid='_871493'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_sdh </ci>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_sdh </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_sdh </ci>
<apply>
<divide/>
<ci> fa </ci>
<ci> Kfa_sdh </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_sdh </ci>
</apply>
<apply>
<divide/>
<ci> fa </ci>
<ci> Kfa_sdh </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_sdh' metaid='_287433' sboTerm='SBO:0000350' units='mmlmin' value='7.38'/>
<parameter id='Ksuc_sdh' metaid='_287435' sboTerm='SBO:0000322' units='mml' value='0.02'/>
<parameter id='Vr_sdh' metaid='_287436' sboTerm='SBO:0000353' units='mmlmin' value='7.31'/>
<parameter id='Kfa_sdh' metaid='_287437' sboTerm='SBO:0000323' units='mml' value='0.4'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='FUM' metaid='_062347'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062347'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R01082'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871505' species='fa'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871517' species='mal'/>
</listOfProducts>
<kineticLaw metaid='_871529'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_fum </ci>
<apply>
<divide/>
<ci> fa </ci>
<ci> Kfa_fum </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_fum </ci>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_fum </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> fa </ci>
<ci> Kfa_fum </ci>
</apply>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_fum </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_fum' metaid='_287438' sboTerm='SBO:0000350' units='mmlmin' value='44.64'/>
<parameter id='Kfa_fum' metaid='_287439' sboTerm='SBO:0000322' units='mml' value='0.15'/>
<parameter id='Vr_fum' metaid='_287440' sboTerm='SBO:0000353' units='mmlmin' value='37.2'/>
<parameter id='Kmal_fum' metaid='_287441' sboTerm='SBO:0000323' units='mml' value='0.04'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='MDH' metaid='_062367'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062367'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00342'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871541' species='mal'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871554' species='oaa'/>
</listOfProducts>
<kineticLaw metaid='_871566'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_mdh </ci>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_mdh </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_mdh </ci>
<apply>
<divide/>
<ci> oaa </ci>
<ci> Koaa_mdh </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_mdh </ci>
</apply>
<apply>
<divide/>
<ci> oaa </ci>
<ci> Koaa_mdh </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_mdh' metaid='_287442' sboTerm='SBO:0000350' units='mmlmin' value='356.64'/>
<parameter id='Kmal_mdh' metaid='_287443' sboTerm='SBO:0000322' units='mml' value='2.6'/>
<parameter id='Vr_mdh' metaid='_287444' sboTerm='SBO:0000353' units='mmlmin' value='353.11'/>
<parameter id='Koaa_mdh' metaid='_287446' sboTerm='SBO:0000323' units='mml' value='0.04'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='ICL' metaid='_062387'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062387'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00479'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871578' species='icit'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871590' species='suc'/>
<speciesReference metaid='_871602' species='gly'/>
</listOfProducts>
<kineticLaw metaid='_871614'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_icl </ci>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icl </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_icl </ci>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_icl </ci>
</apply>
<apply>
<divide/>
<ci> gly </ci>
<ci> Kgly_icl </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icl </ci>
</apply>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_icl </ci>
</apply>
<apply>
<divide/>
<ci> gly </ci>
<ci> Kgly_icl </ci>
</apply>
<apply>
<times/>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icl </ci>
</apply>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_icl </ci>
</apply>
</apply>
<apply>
<times/>
<apply>
<divide/>
<ci> suc </ci>
<ci> Ksuc_icl </ci>
</apply>
<apply>
<divide/>
<ci> gly </ci>
<ci> Kgly_icl </ci>
</apply>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_icl' metaid='_287448' sboTerm='SBO:0000350' units='mmlmin' value='1.9'/>
<parameter id='Kicit_icl' metaid='_287449' sboTerm='SBO:0000322' units='mml' value='0.604'/>
<parameter id='Vr_icl' metaid='_287451' sboTerm='SBO:0000353' units='mmlmin' value='0.019'/>
<parameter id='Ksuc_icl' metaid='_287452' sboTerm='SBO:0000323' units='mml' value='0.59'/>
<parameter id='Kgly_icl' metaid='_287453' sboTerm='SBO:0000323' units='mml' value='0.13'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='MS' metaid='_062407'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062407'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/kegg.reaction/R00472'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871626' species='gly'/>
<speciesReference metaid='_871638' species='aca'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871650' species='mal'/>
<speciesReference metaid='_871662' species='coa'/>
</listOfProducts>
<kineticLaw metaid='_871675'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_ms </ci>
<apply>
<divide/>
<ci> gly </ci>
<ci> Kgly_ms </ci>
</apply>
<apply>
<divide/>
<ci> aca </ci>
<ci> Kaca_ms </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_ms </ci>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_ms </ci>
</apply>
<apply>
<divide/>
<ci> coa </ci>
<ci> Kcoa_ms </ci>
</apply>
</apply>
</apply>
<apply>
<times/>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> gly </ci>
<ci> Kgly_ms </ci>
</apply>
<apply>
<divide/>
<ci> mal </ci>
<ci> Kmal_ms </ci>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> aca </ci>
<ci> Kaca_ms </ci>
</apply>
<apply>
<divide/>
<ci> coa </ci>
<ci> Kcoa_ms </ci>
</apply>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_ms' metaid='_287454' sboTerm='SBO:0000350' units='mmlmin' value='1.9'/>
<parameter id='Kgly_ms' metaid='_287455' sboTerm='SBO:0000322' units='mml' value='2'/>
<parameter id='Kaca_ms' metaid='_287456' sboTerm='SBO:0000322' units='mml' value='0.01'/>
<parameter id='Vr_ms' metaid='_287457' sboTerm='SBO:0000353' units='mmlmin' value='0.019'/>
<parameter id='Kmal_ms' metaid='_287459' sboTerm='SBO:0000323' units='mml' value='1'/>
<parameter id='Kcoa_ms' metaid='_287460' sboTerm='SBO:0000323' units='mml' value='0.1'/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction id='SYN' metaid='_062427'>
<annotation>
<rdf:RDF xmlns:bqbiol='http://biomodels.net/biology-qualifiers/' xmlns:bqmodel='http://biomodels.net/model-qualifiers/' xmlns:rdf='http://www.w3.org/1999/02/22-rdf-syntax-ns#'>
<rdf:Description rdf:about='#_062427'>
<bqbiol:isVersionOf>
<rdf:Bag>
<rdf:li rdf:resource='http://identifiers.org/obo.go/GO:0006103'/>
</rdf:Bag>
</bqbiol:isVersionOf>
</rdf:Description>
</rdf:RDF>
</annotation>
<listOfReactants>
<speciesReference metaid='_871687' species='akg'/>
</listOfReactants>
<listOfProducts>
<speciesReference metaid='_871699' species='biosyn'/>
</listOfProducts>
<listOfModifiers>
<modifierSpeciesReference metaid='_871711' species='icit'/>
</listOfModifiers>
<kineticLaw metaid='_871723'>
<math xmlns='http://www.w3.org/1998/Math/MathML'>
<apply>
<times/>
<ci> cell </ci>
<cn> 0.188 </cn>
<apply>
<divide/>
<apply>
<minus/>
<apply>
<times/>
<ci> Vf_icd </ci>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icd </ci>
</apply>
</apply>
<apply>
<times/>
<ci> Vr_icd </ci>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_icd </ci>
</apply>
</apply>
</apply>
<apply>
<plus/>
<cn> 1 </cn>
<apply>
<divide/>
<ci> icit </ci>
<ci> Kicit_icd </ci>
</apply>
<apply>
<divide/>
<ci> akg </ci>
<ci> Kakg_icd </ci>
</apply>
</apply>
</apply>
</apply>
</math>
<listOfParameters>
<parameter id='Vf_icd' metaid='_287462' sboTerm='SBO:0000350' units='mmlmin' value='14.72'/>
<parameter id='Kicit_icd' metaid='_287464' sboTerm='SBO:0000322' units='mml' value='0.008'/>
<parameter id='Vr_icd' metaid='_287465' sboTerm='SBO:0000353' units='mmlmin' value='0.1472'/>
<parameter id='Kakg_icd' metaid='_287466' sboTerm='SBO:0000323' units='mml' value='0.13'/>
</listOfParameters>
</kineticLaw>
</reaction>
</listOfReactions>
</model>
</sbml>
Simulation of BIOMD0000000222.xml |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Notes |
Original model source: in BioModels database.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | Jarnac 2.14 (http://jdesigner.sourceforge.net/Site/Jarnac.html) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioModels or JWS Online ID | BIOMD0000000222 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Export metadata |
RDF:
metadataModelEntryID44.rdf
XML: metadataModelEntryID44.xml Plain text: metadataModelEntryID44.txt |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Share
|
Cite Model EntryID 44
Cite Model EntryID 44
| APA |
KiMoSys (https://kimosys.org). (2018, December 12). Model EntryID 44 (Escherichia coli). https://kimosys.org/repository/108/associated_models/44
|

|
| MLA |
KiMoSys (https://kimosys.org). "Model EntryID 44 (Escherichia coli)." 2025, kimosys.org/repository/108/associated_models/44.
|

|
| ISO-690 |
KiMoSys (https://kimosys.org). Model EntryID 44 (Escherichia coli). [online], [Accessed 12 December 2025]. Available from: https://kimosys.org/repository/108/associated_models/44
|

|
| BibTeX |
@misc{ KiMoSysModelEntryID44,
author = "{KiMoSys (https://kimosys.org)}",
title = "Model EntryID 44 (Escherichia coli)",
url = "https://kimosys.org/repository/108/associated_models/44",
note = "[Online; accessed 12-December-2025]"
}
|

|
Related Model(s): AccessID 13 | AccessID 16 | AccessID 25 | AccessID 36 | AccessID 38 | AccessID 40 | AccessID 42 | AccessID 43 | AccessID 45
Entered by:
Administrator KiMoSysFirst name: Administrator
Affiliation: INESC-ID/IST
Homepage: http://kdbio.inesc-id.pt/kimosys
Interests: mathematical modeling, accessible data, use of data
Created: 2018-07-23 10:39:44 UTC
Updated: 2018-07-23 10:39:44 UTC
Version: 0
Status: (reviewed) 2018-07-23 10:39:51 UTC
Views: 201
Downloads: 77

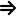 coa + cit
coa + cit